This notebook showcases how to create a workflow using the openEO Python client within the Copernicus Data Space Ecosystem (CDSE). The workflow will be published as a User Defined Process (UDP). UDPs allow you to encapsulate your processing workflow, which consists of multiple steps, into reusable openEO building blocks that can be called with a single command. This approach enables you to share and reuse your workflows across different projects and experiments.
If your workflow is part of a scientific or research experiment, you can publish it in the EarthCODE Open Science Data Catalog once finalized. This ensures that your workflow is findable and accessible to other users in the scientific community.
In this example, we will create a workflow to generate a variability map, using Sentinel-2 data products that are available on the Copernicus Data Space Ecosystem. . In a variability map, each image pixel is assigned a specific category, which represents the deviation of the pixel value from the mean pixel value. These maps can be used to implement precision agriculture practices by applying different fertilization strategies for each category. For instance, a farmer might choose to apply more product to areas of the field that exhibit a negative deviation and less to those with positive deviations (enhancing poorer areas), or concentrate the application on regions with positive deviations (focusing on the more productive areas of the field).
import rasterio
import matplotlib.pyplot as plt
def visualise_tif(path: str):
with rasterio.open(path) as src:
data = src.read(1) # Read the first band
plt.figure(figsize=(10, 10))
plt.imshow(data, cmap='viridis')
plt.colorbar()
plt.show()Connection with CDSE openEO Federation¶
The first step, before creating any processing workflow in openEO, is to authenticate with an available openEO backend. In this example, we will use the CDSE openEO federation, which provides seamless access to both datasets and processing resources across multiple federated openEO backends.
import openeo
import json
from openeo.rest.udp import build_process_dictconnection = openeo.connect(url="openeofed.dataspace.copernicus.eu").authenticate_oidc()Authenticated using refresh token.
Defining the workflow parameters¶
The first step in creating an openEO workflow is specifying the input parameters. These parameters enable users to execute the workflow with their own custom settings, making it adaptable to different datasets and use cases. openEO provides built-in helper functions that assist in defining these parameters correctly.
from openeo.api.process import Parameterarea_of_interest = Parameter.geojson(name='spatial_extent', description="Spatial extent for which to generate the variability map")
time_of_interest = Parameter.date(name='date', description="Date for which to generate the variability map")Implementation of the workflow¶
Next, we will begin implementing the variability map workflow. This involves using the predefined functions in openEO to create a straightforward workflow consisting of the following steps:
Select the S2 data based on the
area_of_interestandtime_of_interestparameters.Calculate the NDVI for the S2 data.
Apply an openEO User Defined Function (UDF) to calculate the deviation of each pixel against the mean pixel value of the datacube.
# Step 1. Select the S2 data based on the workflow parameters
s2_cube = connection.load_collection(
"SENTINEL2_L2A",
spatial_extent=area_of_interest,
temporal_extent=[time_of_interest,time_of_interest],
)
s2_masked = s2_cube.mask_polygon(area_of_interest)# Step 2. Calculate the S2 NDVI
s2_ndvi = s2_masked.ndvi()# Step 3. Apply the UDF to calculate the variability map
calculate_udf = openeo.UDF.from_file("./files/variability_map.py")
varmap_dc = s2_ndvi.reduce_temporal(calculate_udf)from IPython.display import JSON
JSON(varmap_dc.to_json())Create an openEO-based workflow¶
In this next step, we will create our workflow by establishing our openEO User Defined Process (UDP). This action will create a public reference to the workflow we developed in the preceding steps. This can be achieved by using the save_user_defined_process function.
workflow = connection.save_user_defined_process(
"variability_map",
varmap_dc,
parameters=[area_of_interest, time_of_interest],
public=True
)
workflowIn the previous step, we created a workflow as a UDP in openEO. We can now use the public URL to share the workflow with others or to execute it in different contexts. The UDP encapsulates the entire processing logic, making it easy to apply the same workflow to different datasets or parameters without needing to redefine the steps each time. In this example, the published UDP is available at the following URL: Variability Map UDP.
Testing the workflow¶
After saving the workflow, we can test it by executing the UDP with specific parameters. This step allows us to verify that the workflow operates as expected and produces the desired results. We start by defining the parameters that we want to use for the test run. These parameters will be passed to the UDP when it is executed.
spatial_extent_value = {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {},
"geometry": {
"coordinates": [
[
[
5.170043941798298,
51.25050990858725
],
[
5.171035037521989,
51.24865722468999
],
[
5.178521828188366,
51.24674578027137
],
[
5.179084341977159,
51.24984764553983
],
[
5.170043941798298,
51.25050990858725
]
]
],
"type": "Polygon"
}
}
]
}
date_value = "2025-05-01"Next we use our previously created datacube varmap_dc to execute our workflow as an openEO batch job. This step involves submitting the job to the openEO backend, which will process the data according to the defined workflow and parameters. The backend will handle the execution of the workflow and return the results, which can then be analyzed or visualized as needed.
path = "./files/varmap_workflow_test.tiff"
varmap_test = connection.datacube_from_process(
"variability_map",
spatial_extent=spatial_extent_value,
date=date_value,
)
varmap_test.execute_batch(
path,
title="CDSE Federation - Variability Map Workflow Test",
description="This is an example of a workflow test containing the calculation of a variability map in Belgium",
)Finally, we can visualize the results of our workflow. This step allows us to see the output of the variability map and assess its quality and relevance for our specific use case. Visualization is a crucial part of the workflow, as it helps in interpreting the results and making informed decisions based on the data processed by our openEO workflow.
visualise_tif(path)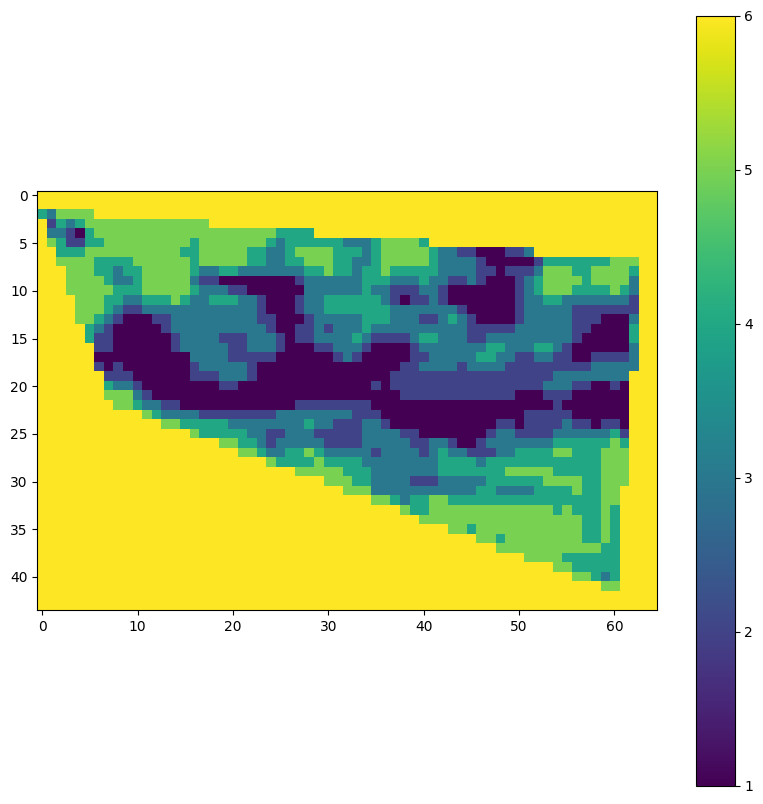
Sharing your workflow¶
Now that our workflow has been created using save_user_defined_process and we’ve confirmed that it works, we can share it with others and the broader community. Using the openEO functions demonstrated before, the workflow is automatically stored on the openEO backend we connected to in the initial steps. The workflow, referred to as a User Defined Process (UDP) in openEO terminology, is a JSON-based structure that contains the steps of the workflow, represented as an openEO process graph, along with additional metadata such as a description of the workflow parameters.
Exporting your workflow¶
There are several ways to make your workflow accessible for reuse among peers and within your communities:
Share the public URL with your peers
Since we usedpublic=Truein oursave_user_defined_process, a public URL was automatically added to the workflow definition. In this case, the public URL is:https://openeo .dataspace .copernicus .eu /openeo /1 .1 /processes /u:6391851f -9042 -4108 -8b2a -3dd2e8a9dd0b /variability _map Export the workflow definition to your preferred storage
Alternatively, you can also export the workflow and store it in a version-controlled environment like GitHub or your own preferred storage. This gives you full control over its content and version history. In this case, instead of usingsave_user_defined_process, you can usebuild_process_dictto create a dictionary representation of the workflow, which can then be written to a file. However, if you want others to reuse your workflow, make sure the file is accessible via a public URL. This is necessary for the openEO backend to retrieve and execute the workflow definition.
spec = build_process_dict(
process_id="variability_map",
process_graph=varmap_dc,
parameters=[area_of_interest, time_of_interest],
)
with open("files/variability_map_workflow.json", "w") as f:
json.dump(spec, f, indent=2)Sharing your workflow¶
Once you have a public reference to your workflow, either through the openEO backend or a public URL pointing to a definition stored on GitHub or another platform, you can share it in various ways to enable others to execute it, as shown in our Creating an experiment example. There are many different ways to share your workflow:
Direct sharing with peers and communities
Publishing your workflow on the EarthCODE Open Science Catalogue, as demonstrated in our publication example
Publishing on platform marketplaces, such as the Copernicus Data Space Ecosystem Algorithm Plaza.