System requirements¶
- Ubuntu OS min version > 20. Script tested on Ubuntu 22.04 and Ubuntu 20.04 version.
- Minimum requirements: 2 GB RAM, 2 CPUs, 32 GB of disk space.
- User with full sudo privileges to run all Linux commands and install packages as root.
- Selection of packages to be installed in order to run bash script: python 3 , gdal , tree, jq, parallel , curl, stactools.
- Python 3.8 or greater
- Following dependencies are necessary to make correct conversion of products to be published:
stactools- to manage STAC catalogsstactools-datacube- to enrich STAC items with datacube metadata Please find full documentation and installation instructions here: https://stactools .readthedocs .io /en /stable /index .html
Add metadata of a single product (item) to the catalogue¶
Manual ingestion of single item into catalogue is rather simple and straightforward when you have all metadata prepared correctly and when you follow these few steps on how to add new product (collection.json) to a publicly open repository of Open Science Data Catalogue.
Please note that this workflow is applicable also to other elements of the catalogue such as Projects, Themes, Variables, EO Missions. Here the procedure of adding or updating metadata of single item using GitHub on Web browser is provided.
- Go to open-science-catalog-metadata-staging repository:
https://
github .com /EOEPCA /open -science -catalog -metadata -staging
- Go to /products/ folder to extend the list of products:
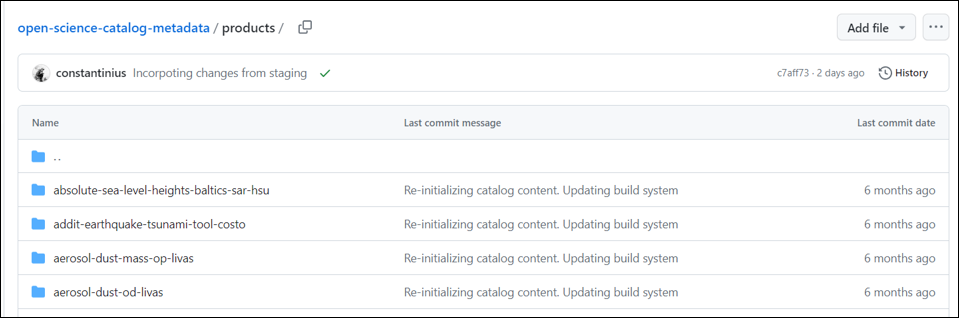
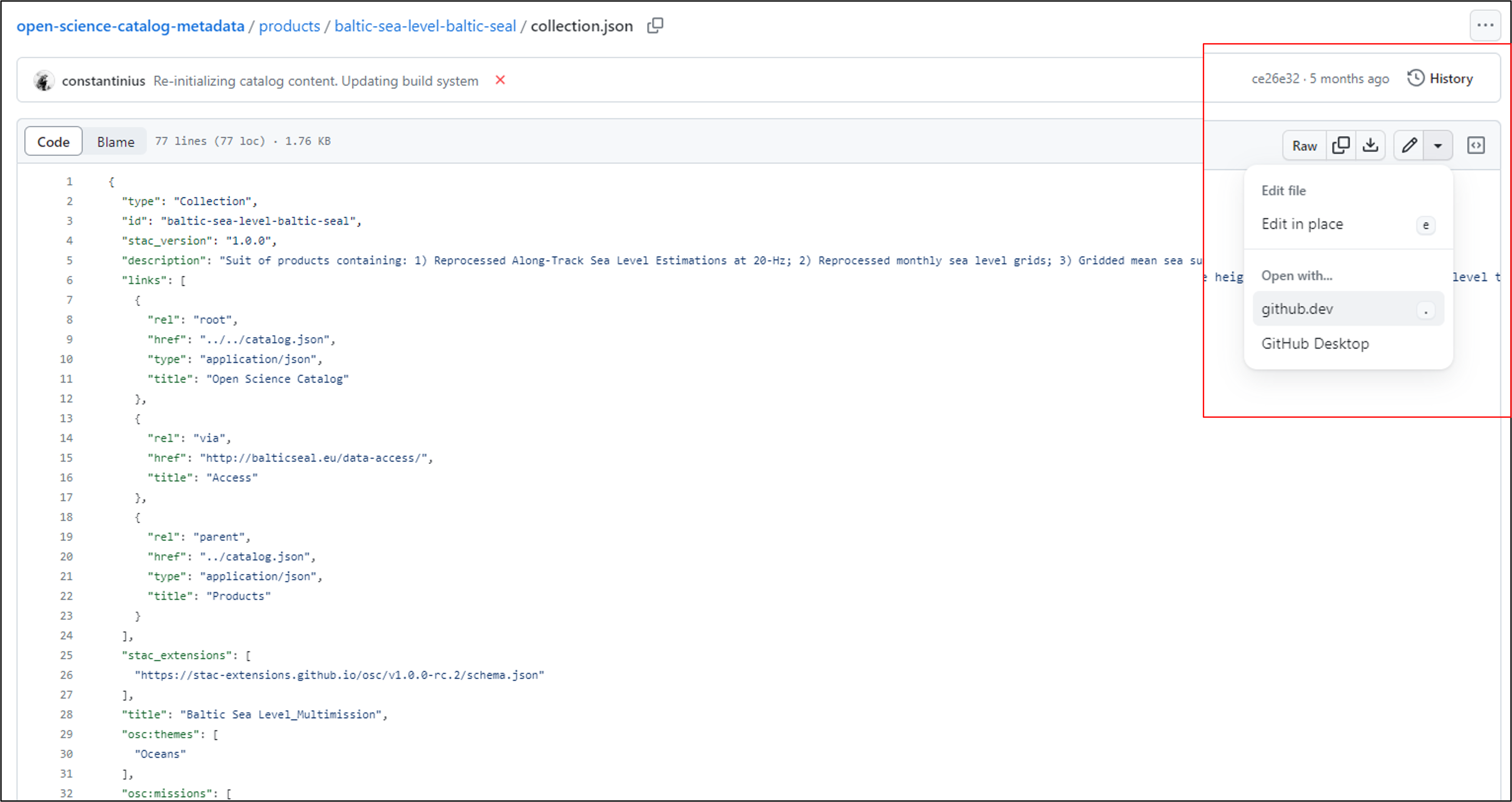
Open any folder with the short product name and check the metadata file stored in collection.json format.
Click on Editing mode and open JSON file with github.dev to make changes or to copy the content of JSON file in web-based editor.
- In GutHub web-based text editor (github.dev), select “Source controller” and “Create a new branch”
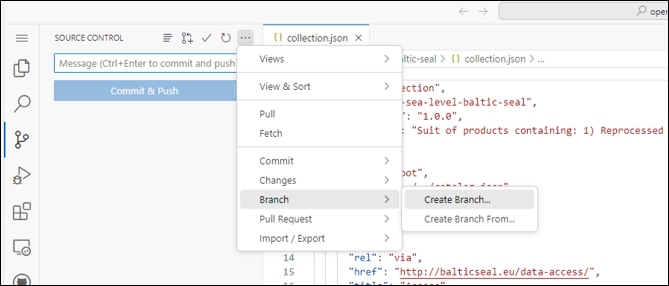
- Switch to new Branch:
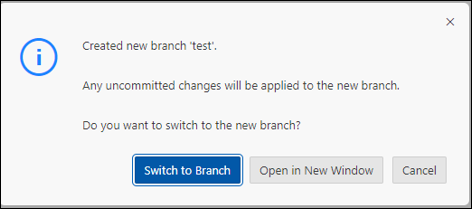
Go to Explorer and Add a new folder where you can store the collection.json with metadata of your Product. Name your folder with a unique name (id) that you give to your product!
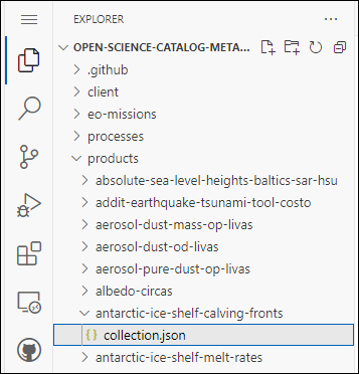
Create collection.json file and add all metadata needed for the product. The simplest way to create a new collection.json is to CTRL+A and CTRL+C of existing collection.json (even from different project and CTRL+V to a new empty collection.json created.
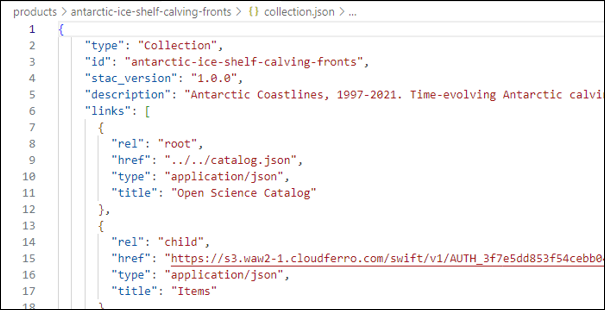
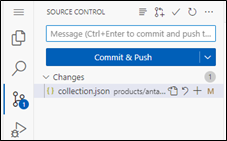
- You can see the file being modified with the capital letter M.

- Commit and Push changes from Source Control Panel. Write a purpose or a subject of changes made in the “Message” field.
- Create Pull Request to request changes in the repository!
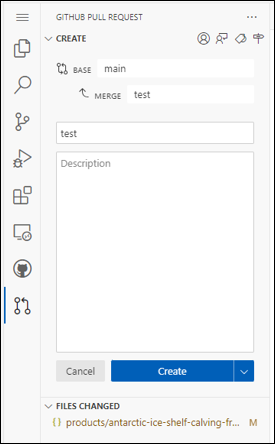
- Once created the Pull Request will be accepted or rejected by open-science-catalog-metadata administrator.
Ingest metadata of assets with STAC Catalog¶
As mentioned in the previous sub-chapter metadata repository is stored in the GitHub repository. For ingestion of data that are stored in structural catalog, user should convert this file structure to ingest to STAC catalog. Then created STAC collection can be directly imported to GitHub and merged to existing Open Science Data Catalogue repository.
To convert the file structure to STAC collection, please refer to stactools documentation, which describes best practices on creation of such collection.
Data owners interested in ingestion of multiple products to the catalog are asked to convert their dataset directly into STAC items collection. This especially refers to large datasets (e.g. multitemporal .nc files containing monthly products for a time period of 20 years). Is such case data owners or principal investigators of the project will be provided with script which facilitates that process of ingestions if such products, allowing their conversion to .json STAC items collection.
Add multiple or single product(s) stored in external server (open-access storage) with STAC Catalogue¶
In the previous section the steps applied described a process to ingest metadata and the access to products stored in external server which may provide open data access to anyone or to registered users in some cases.
In this scenario complete instructions are provided for how to import larger datasets when multiple changes to the files stored in open science data catalog in GitHub repository are required. With our current system, it is impossible for the user to keep track 10+ open pull requests, so when user requires multiple changes at once, Git with a code editor is recommended.
The description on how to import STAC catalogue which is preferred method of an ingestion of asset metadata is described in details. This will allow end users to access metadata of individual granules (stored originally in the cloud-native external server) and download them to local storage separately without the need of bulk downloading of all files at once. The workflow includes:
Find the description of the dataset in Open Science Data Catalogue frontend. If the product (dataset) does not exist in the catalog yet, add it to the catalog first! (see subchapter above).
Prepare an upload dataset (assets / granules) This step is a prerequisite that must be met for any data collection to make it publicly available and allow end user to discover and use the data. This can be done in two modes: a) uploading data to s3 bucket administrated by OSC developers, b) provide link to capable HTTP where files are already available to download or / and preview.
In this case assets are stored in open-access repository: Maddalena et al. (2023).
Therefore, no preliminary steps are required related to data relocation from this source repository. The preparation of the dataset must consist first of assessing the exact full path to original repository for each granule. In this case the list of products is loaded and save in standard text file:
curl -sL "https://zenodo.org/record/7568049#.ZDbG4nbP1aQ" | grep -oP '<a href="\K([^"]+\.tif)' > tiff_links.txt ; sed -i 's|^|https://zenodo.org|' tiff_links.txt- In case of a long list of files to be converted at once it is advised to split the list of links for each granule into smaller list. It has been confirmed that up to 10 files can be written to JSON format at once using the tool proposed.
- Create STAC Catalog for the dataset
a. Create a structure of catalog.json (could only be 1, but depending on the dataset size and structure it can be more)
b. Create STAC Items data items (granules). Either:
i. for 2D raster datasets: _ stac create-item _
ii. for netcdfs/ZARRs stac datacube create
iii. other tools?
iv. or manually***
In this scenario a 2D raster datasets are used and the STAC catalog is created with open-source tool ‘stac create-item’ (i). In case of multiple items to be converted at once, a for loop is created to create stac item .json file for each granule by accessing separate item in a loop:
mkdir item_files ; for line in $(cat tiff_links.txt); do item_json="item_files/item_$line_number.json"; echo $line ; stac create-item "$line" | tee "$item_json"; sleep 3; ((line_number++)); done < "tiff_links.txt"** Note: once files are created, you can access the metadata by opening and reading single item.json file with cat item.json
*** Future updates in the guide are foreseen and guidance specific to most commonly used data formats will be provided. To manually create JSON file, please refer to general file structure provided in the STAC documentation: examples
c. Add STAC Items into Catalog structure
To create appropriately catalog.json file that lists all related items to specific collection and gathers them in the catalog, it is necessary to follow strict catalog.json file formatting as suggested in:
https://github.com/radiantearth/stac-spec/blob/master/examples/catalog.json
Please download the template on how this catalog.json should looks like and upload it into the directory where the item files are stored.
Make necessary correction to imported catalog.json example file:
the only changes which are required is to remove “child” and “item” entries from the catalog.json.
Item entries will be updated automatically once you add the assets.
Update the default description and title of the catalog
The final catalog.json should looks like this:
{
"type": "Catalog",
"id": "examples",
"title": "Example catalog",
"stac_version": "1.0.0",
"description": "This catalog is a simple demonstration of an example catalog that is used to organize STAC Items",
"links": [
{
"rel": "self",
"href": "https://raw.githubusercontent.com/radiantearth/stac-spec/v1.1.0/examples/catalog.json",
"type": "application/json"
},
{
"rel": "root",
"href": "./catalog.json",
"type": "application/json",
"title": "Example catalog"
}
]
}NOTE: While working on Linux based environment you can make changes with vi catalog.json in terminal.
- **Add STAC Items to a common _catalog.json _ by applying _ ‘stac add’ _ command
for item_file in item_files/item_*.json; do stac add "$item_file" catalog.json; done;- Export the catalog structure _
stac copy_
Before performing this step, remember to change the directory from the input data collection to output repository which will be copied to s3 bucket!
stac copy catalog.json item_10files/out_json/ -l[https://s3.waw2-1.cloudferro.com/swift/v1/AUTH_3f7e5dd853f54cebb046a29a69f1bba6/Catalogs/4DGreenland/supraglacial-storage-and-drainage-lake-features-mapped-by-sentinel1/catalog.json](https://s3.waw2-1.cloudferro.com/swift/v1/AUTH_3f7e5dd853f54cebb046a29a69f1bba6/Catalogs/4DGreenland/supraglacial-storage-and-drainage-lake-features-mapped-by-sentinel1/catalog.json)NOTE: Check first the location of your product within the Open Science Data Catalog repository!
- Upload STAC Catalog to S3 or another HTTP service
With this simple command all JSON files (single granules) are moved to dedicated OSC s3 bucket metadata repository:
s3cmd sync item_10files/out_json/ s3://Catalogs/4DGreenland/supraglacial-storage-and-drainage-lake-features-mapped-by-sentinel1/
- Add reference to Product’s metadata and give access to created STAC Catalog
In OSC Catalogue find target product which needs to be updated with the STAC Item’s collection. To perform this step 'href’ link must be updated, by inserting the list to catalog.json file stored in s3 repository to Product’s collection.json file. As shown below:
{
"rel": "child",
"href": "https://s3.waw2-1.cloudferro.com/swift/v1/AUTH_3f7e5dd853f54cebb046a29a69f1bba6/Catalogs/4DGreenland/supraglacial-storage-and-drainage-lake-features-mapped-by-sentinel1/catalog.json",
"type": "application/json",
"title": "Items"
}NOTE: You can make changes using GitHub Desktop or within web-based editor as shown in the section before.
- Commit changes and create Pull Request
Changes in Product description will be reviewed by OSC administrator and accepted or rejected by administrator.
- Preview uploaded STAC granules collection in the Open Science Data Catalogue frontend:
Add multiple or single product(s) originally stored in local repository with STAC Catalogue¶
In this scenario large open-source datacube was investigated and made available to be used by open public. Steps presented in this scenario will allow to first change the location of the dataset to make them accessible to all users and create STAC Catalog filled with metadata from datacube. This will allow end users to access metadata of datacube which is accessible, downloadable and can be visualized or further processed in other locations (e.g. Jupyter Notebooks).
- Prepare an upload data (assets / granules)
This step is a prerequisite that must be met for any data collection to make it publicly available and allow end user to discover and use the data. This can be done in two modes:
1) uploading data to s3 bucket administrated by OSC developers, 2) provide link to capable HTTP where files are already available to download or / and preview.
In this case the first mode will be used. Datacube is stored in .zip archive making it impossible to be directly accessed and explored. Therefore, a preliminary step required related to data relocation from source repository: Alonso et al. (2023)to a dedicated folder in OSCAssets repository in Open Science Data Catalogue S3 storage.
File relocation requires configured access to dedicated S3 bucket which in this case is granted only to authorized contributors.
Once configured, downloaded product should be extracted and relocated into designated storage, where it will become accessible for anyone.
s3cmd sync SeasFireCube_v3.zarr s3://OSCAssets/seasfire/seasfire-cube/SeasFireCube_v3.zarr- Create a STAC Catalog for the data
a. Create a structure of catalog.json (could only be 1 if not more is necessary)
b. Create STAC Items data items (granules). Either:
i. for 2D raster datasets_:
stac create-item_ ii. for netcdfs/ZARRs _stac datacube create-item_ iii. other tools? iv. or manually
** Future updates in the guide are foreseen and guidance specific to most commonly used data formats will be provided. To manually create catalog.json please refer to basic file structure provided in the STAC documentation.
In this scenario a 3D datacube is used and the STAC catalog is created with open-source tool ‘stac datacube create-item’. This command uses ‘stactools’ extension package which allows to create or extend STAC Items dealing with multi-dimensional data formats and to extract datacube related metadata from these assets. Full documentation and the latest release of the package can be found under the link here: https://
stac datacube create-item s3://OSCAssets/seasfire/seasfire-cube/SeasFireCube_v3.zarr/ item.json '--use-driver ZARR** Note: once files are created, you can access the metadata by opening and reading single item.json file with cat item.json
Next steps taken to enable product to be discoverable within STAC Catalog are the same as in previous use case (see point 2c - 5) and follow the steps described in this previous section.
** Remember to change the folder name and product folder in s3 bucket to relate to adequate product in OSC**
Commit changes and create Pull Request Preview uploaded STAC Item catalog: Changes in Product description will be reviewed by OSC administrator and accepted or rejected by administrator.
Preview uploaded STAC granules collection in the Open Science Data Catalogue frontend: https://
opensciencedata .esa .int /products /seasfire -cube /collection Data can be visualized using Jupyter Notebook and dedicated ‘xarray’ Python package to further work with the datacube on-the-cloud and visualize variables.
Full metadata contained in the .zarr file can also be read directly from the browser under: https://
Add multiple assets at once with GitHub¶
Once created the JSON files describing each single asset of larger products collection can be imported to Open Science Data Catalogue repository of metadata at once using GitHub.
For this, GitHub CLI or gh should be used. GitHub CLI is a command-line interface to GitHub for use in terminal or scripts. It facilitates the process of making changes in open access github repository as the one at open-science-data-catalog-metadata and allows to ingest several files at once. To work with this command-line tool. Please check the installation steps first: docs
To correctly install gh tool, follow these instructions. Please note that instructions provided here refer to Linux Ubuntu OS, and have not been tested on any other software:
- Install System dependencies:
sudo apt update
sudo apt install -y git python3 python3-pip gdal-bin tree jq parallel curl- Install ‘gh’ tool. In case you encounter any issues please follow instructions from here: docs
/install _linux .md
type -p curl \>/dev/null || (sudo apt update && sudo apt install curl -y)
curl -fsSL https://cli.github.com/packages/githubcli-archive-keyring.gpg | sudo dd of=/usr/share/keyrings/githubcli-archive-keyring.gpg \
&& sudo chmod go+r /usr/share/keyrings/githubcli-archive-keyring.gpg \
&& echo "deb [arch=$(dpkg --print-architecture) signed-by=/usr/share/keyrings/githubcli-archive-keyring.gpg] https://cli.github.com/packages stable main" | sudo tee /etc/apt/sources.list.d/github-cli.list \> /dev/null \
&& sudo apt update \
&& sudo apt install gh -y- Install Python dependencies
Following dependencies are necessary to make correct conversion of products to be published:
stactools- to manage STAC catalogsstactools-datacube- to enrich STAC items with datacube metadata
To install following packages copy and paste following in your terminal:
pip install -U stactools stactools-datacube4.Ingest products to GitHub metadata repository using GitHub CLI
# clone the git repository:
gh repo clone \<insert-user-here\>/open-science-catalog-metadata.git
# enter repository
cd open-science-catalog-metadata
# create a new branch to work on
git checkout -b \<branch-name\>
cd ..
# merge the output catalog to the metadata repository
stac merge --as-child \
\<folder-with-JSON-files/catalog.json \
# go to the repo again and commit all new/changed files
cd open-science-catalog-metadata
git add \<project-name\>/\<product-name\>
git commit -m"Adding woc-l4-se-erastar-h\_v2.0"
git push --set-upstream origin \<branch-name\>
gh pr create -f- Check the status of Pull Requests in GitHub:
https://
- Changes to the Catalogue content will be reviewed and accepted or rejected by the OSC Administrator.
- Maddalena, J., Leeson A, & McMillan M. (2023). Supraglacial lakes derived from Sentinel-1 SAR imagery over the Watson basin on the Greenland Ice Sheet. Zenodo. 10.5281/ZENODO.7568049
- Alonso, L., Gans, F., Karasante, I., Ahuja, A., Prapas, I., Kondylatos, S., Papoutsis, I., Panagiotou, E., Mihail, D., Cremer, F., Weber, U., & Carvalhais, N. (2023). SeasFire Cube: A Global Dataset for Seasonal Fire Modeling in the Earth System. Zenodo. 10.5281/ZENODO.8055879